"Celery Event Reporting: Getting Your Jobs on that Hotline Bling."
There are two things we can all agree on: 1. Drake dropped a massive hit with his single “Hotline Bling” and 2. celery is a great distributed computing library for a wide range of needs. That being said, celery (much like Drake’s complex relationship status as described in his song) is not for the faint of heart. celery can feel too configurable: it has all sorts of bells and whistles, but sometimes it can be difficult to know how to get it to do what you want. One necessary use case is reporting: what your are jobs doing and what celery is doing with them. The default settings aren’t great for this so if you don’t set things up correctly, just like Drake’s lament about his erstwhile girlfriend, you will be in a state of wondering what’s going on. So let’s get them on that hotline bling.
Here we just want to accomplish two things:
1. Set some reasonable configuration settings for a basic use case of distributed task execution
2. Monitor celery running live to verify that it is performing as desired
For my case, I want to execute Numba-driven Python calculations that take quite a bit of number crunching. Each job has the following characteristics:
1. significant memory high watermark
2. >= 20 seconds run time
3. CPU-bound for most of the time
It turns out that celery does certain things by default that are not good for these kinds of jobs. In particular, celery workers by default will attempt to grab a few jobs from the queue at once, instead of just grabbing one job at a time. This may seem odd, but the idea is that the overhead of going to the queue and asking for jobs is then amortized over several job executions. For my case though, job execution time dominates over any latency of going to the queue, so I want to turn off this behavior. “Turning off” the prefetch behavior is really the same thing as setting the prefetch setting to 1, so you have to give celery this setting when you start up:
CELERY_PREFETCH_MULTIPLIER=1 celery -A celery_tasks worker -l infoWhat about concurrency? The celery docs say that the default setting here is to set the number of workers to be the same as the number of CPUs reported by the OS. For my jobs, I’m mostly CPU-bound, so that’s a reasonable setting. If your jobs are often sitting around waiting for network traffic or the like, you might want to set that number higher. The setting is CELERYD_CONCURRENCY.
But now I want to load test and prove that celery is doing what I want (prefetch setting to 1 and max concurrent jobs set to number of CPUs). If I get this wrong in production, too many jobs will launch and I’ll get memory exceptions (due to my high memory watermark). Much like Drake, my level of trust here is low. In the documentation for event reporting, you will find all sorts of interesting ways to tweak the reporting of events, but you won’t find the actual magic command you have to enter so that you can actually see what your jobs are doing.
The trick is to tell celery that you would like to enable events, and then it will allow you to view tasks as they come in with the events command:
$ celery -A celery_tasks control enable_events
$ celery -A celery_tasks eventsThis will bring up a screen that will show you exactly what’s going on as jobs are run on your system:
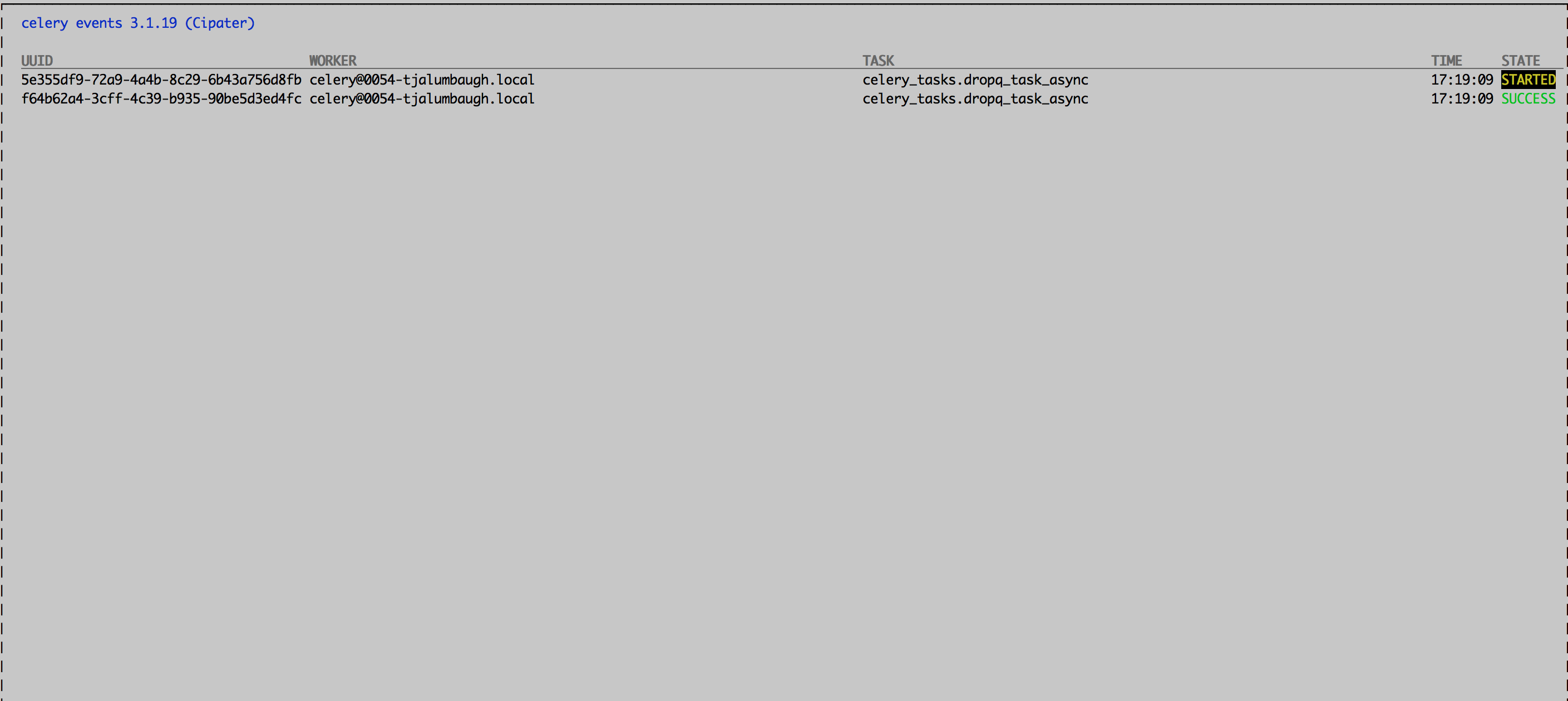
What about when your jobs complete? Celery provides a number of mechanisms for you to figure out that those jobs you thought were so well-behaved were actually living a wild wild nightlife. It turns out that my original way of handling generated celery tasks works, but only for certain cases. So here is what I originally did:
Step 1: After launching an asynchronous task, just return its job ID and use that as your handle for the job.
handle = my_task.delay(arg1, arg2)
return str(handle)Step 2. At some later time, create a fresh AsyncResult object from that job ID.
# Use the job_id to create an AsyncResult and get its status
result = celery_app.AsyncResult(job_id)
if result.ready():
return resultsresultThe worst thing about this solution is that it works some of the time. This result object will respond correctly to ready() as long as the only thing that ever happens to your job is that it executes succesfully. If your job does anything other than succeed, this technique will always tell you that your job is in the PENDING state. Forever. With no error message. So, I gathered that I shouldn’t do that.
Instead, you should keep track of the actual object returned by the delay method; here I use a dictionary:
job_handle = my_task.delay(arg1, arg2)
# Keep the handle!
running_jobs[job_handle.id] = job_handleThis object will hold the proper status of the task (e.g. ‘SUCCESS’, ‘FAILURE’, ‘PENDING’, etc.) and even give you the traceback if an exception is raised:
# Now it's time to check if the job is done
task = running_jobs[job_id]
if task.ready():
if task.successful():
return results.result
elif results.failed():
return results.traceback
else:
# Nothing should get here
raiseSo, with those two techniques (enabling/viewing events and keeping track of generated AsyncResults objects) you’ll stay in touch with all of your celery jobs and know exactly what they are up to.
This will likely lower your angst level about your celery jobs and generally help you avoid having to do things like this: